Hippocampus Illumina NOS (Oct08) RankInv beta

Accession number: GN213
Summary:
EXPERIMENTAL DATA SET (Unpublished): This is one of five INIA companion data sets generated using the hippocampus of LXS strains and the Illumina Mouse 6.1 bead array. The data set labeled Hippocampus Illumina (May07) RankInv provides baseline control expression data with no treatment at all. This NOS data set consists of animals who received a single IP injection of saline (NOS = no restraint saline) without restraint stress. This saline injection group is intended to provide appropriate control for cases that received an IP injection of ethanol. The only experimental stressor in this NOS data set is that associated with handling and the IP saline injection. Survival period was 4 hours. The paradigm that was used in this set of studies by Lu Lu and colleagues is identical to that used by Dr. Michael Miles (see his experimental prefrontal cortex data in GeneNetwork for both LXS and BXD strains).
The hippocampus is highly susceptible to the effects of stress and glucocorticoid hormone action. ADD TEXT, rationale, and links.
Samples were processed using a total of 12 Illumina Sentrix Mouse 6.1 Bead arrays.
About the strains used to generate this set of data:
The LXS genetic reference panel of recombinant inbred strains consists of just over 70 strains. The LXS strains in this data set were obtained from Dr. Beth Bennett and colleagues at the University of Colorado, Bolder. All of these strains are fully inbred, many well beyond the 25th filial (F) generation of inbreeding. All of these LXS strains have been genotyped at 13,377 SNPs.
About the animals and tissue used to generate this set of data:
All animals were raised at the University of Colorado or at University of Memphis in SPF facilities. All mice were killed by cervical dislocation. Whole brain dissections were performed at UTHSC by Lu Lu and colleagues. Hippocampal samples were close to complete but are likely to include variable amounts of fimbira and choroid plexus. Samples may also include parts of the subiculum.
The bilateral hippocampus tissue from one naive adults mouse was used to generate RNA samples. All animals were sacrificed between 9 AM and 5 PM during the light phase. All RNA samples were extracted at UTHSC by Feng Jiao.
All animals used in this study were between 60 and 74 days of age (average of 67 days; see Table 1 below). All animals were sacrifice between 9 AM and 5 PM during the light phase.
Sample Processing: Samples were processed by Lu Lu and colleagues in the Illumina Core at UTHSC between April 2007 and November 2007. All processing steps were performed by Feng Jiao. In brief, RNA purity was evaluated using the 260/280 nm absorbance ratio, and values had to be greater than 1.8 to pass our quality control (QC). The majority of samples had values between 1.9 and 2.1. RNA integrity was assessed using the Agilent Bioanalyzer 2100. The standard Eberwine T7 polymerase method was used to catalyze the synthesis of cDNA template from polyA-tailed RNA using the Ambion/Illumina (http://www.ambion.com/catalog/CatNum.php?AMIL1791). TotalPrep RNA amplication kit (Cat#IL1791). The biotin labeled cRNA was then evaluated using both the 260/280 ratio (values of 2.0-2.3 are acceptable) using a NanoDrop ND-1000 (http://www.nanodrop.com/nd-1000-overview.html). Those samples that passed QC steps (1-3% failed and new RNA samples had to be acquired and processed) were immediately used on Mouse-6 v 1.1 slide. The slides were hybridized and washed following standard Illumina protocols.
Replication and Sample Balance: We obtained matched male and female sample from 24 strains. The following seven strains are represented by male samples only: ILS, LXS25, 39, 43, 54, 78, and 110. Two strains, LXS19 and 92, are represented only by female samples.
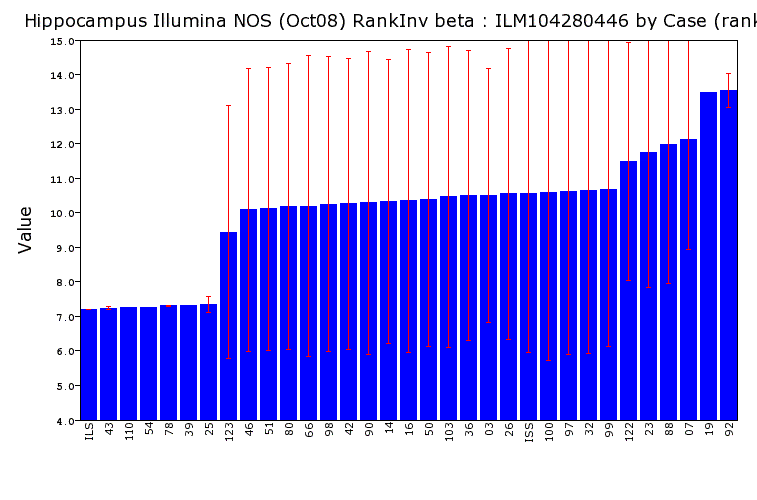
Legend:Sex balance of the NOS data set is revealed by expression of the Xist RNA (Illumina probe ILM1042800446). Male samples have low expression of Xist (about 7 units), whereas females have high expression (about 13 units). Each bar provides the mean expression value (log2 transformed) for a single strain. Strains with both male and female samples have intermediate averages and large error bars.
Experimental Design and Batch Structure: This data set consists arrays processed in 17 groups over a seven month period (from April 2007 to November 2007). Most groups consisted of 5 samples. All arrays in this data set were processed using a single protocol by a single operator, Feng Jiao. Processing was supervised directly by Dr. Lu Lu. All samples were scanned on a single Illumina Beadstation housed in the Hamilton Eye Institute between April 24, 2007 and November 20, 2007 . Details on sample assignment to slides and batches is provide in the table below.
Error checking
- Checked for genotypes of LXS strains using a battery of test Mendelian transcripts (transcripts with a Mendelian segregation pattern in the LXS strains). Peak LRS of 201.7 for C1orf57 using Illumina probe ILM110129. There are no known errors in the strain assignment. (NOS data set)
These genotype discrepancies are either due to recombination between the marker and the probe or a genotyping errors. (RWW, Feb 27, 2008)
- Total count of transcripts/probes with LRS greater than 46 (LOD>10) is 754 with 31 LXS strains (NOS data set).
NOS DATA AS OF Oct 2008: Data Table 1:
This table lists all arrays by order of strain (index) and includes data on tube ID, strain, age, sex, F generation number, number of animals in each sample pool (pool size), slide ID, slide position (A through F), scan date, and scan batch.
| Index | Tube ID | Strain | Age | Sex | Pool Size | Slide ID | Slide position | Batch by slide | Scan batch |
| 1 | R3731H | ILS | 72 | M | 1 | 1714451029 | A | 1 | 7 |
| 2 | R3730H | ILS | 72 | M | 1 | 1714451039 | A | 2 | 6 |
| 3 | R3933H | ISS | 64 | F | 1 | 1735640068 | D | 8 | 14 |
| 4 | R3885H | ISS | 66 | M | 1 | 1736925322 | F | 19 | 9 |
| 5 | R3804H | LXS3 | 65 | M | 1 | 1825397042 | B | 27 | 8 |
| 6 | R3942H | LXS3 | 68 | F | 1 | 1825397078 | F | 32 | 12 |
| 7 | R3851H | LXS3 | 65 | F | 1 | 1833451001 | A | 38 | 17 |
| 8 | R3852H | LXS3 | 63 | M | 1 | 1833451010 | B | 40 | 19 |
| 9 | R3769H | LXS7 | 73 | F | 1 | 1825397036 | D | 25 | 15 |
| 10 | R3841H | LXS7 | 60 | F | 1 | 1825397080 | D | 33 | 16 |
| 11 | R3770H | LXS7 | 73 | F | 1 | 1825397100 | C | 35 | 17 |
| 12 | R3825H | LXS7 | 66 | M | 1 | 1870382102 | B | 52 | 13 |
| 13 | R3727H | LXS14 | 70 | M | 1 | 1825397035 | F | 24 | 15 |
| 14 | R3715H | LXS14 | 70 | F | 1 | 1825397076 | D | 30 | 12 |
| 15 | R3902H | LXS16 | 72 | F | 1 | 1736925146 | F | 12 | 9 |
| 16 | R3916H | LXS16 | 73 | M | 1 | 1825397078 | B | 32 | 12 |
| 17 | R3862H | LXS19 | 67 | F | 1 | 1736925148 | A | 13 | 10 |
| 18 | R3680H | LXS23 | 65 | F | 1 | 1736925131 | C | 11 | 4 |
| 19 | R3681H | LXS23 | 64 | M | 1 | 1736925131 | D | 11 | 4 |
| 20 | R3803H | LXS23 | 65 | F | 1 | 1736925148 | B | 13 | 10 |
| 21 | R3754H | LXS25 | 70 | M | 1 | 1825397076 | E | 30 | 12 |
| 22 | R3753H | LXS25 | 70 | M | 1 | 1848071016 | D | 57 | 19 |
| 23 | R3835H | LXS26 | 64 | F | 1 | 1825397081 | F | 34 | 16 |
| 24 | R3836H | LXS26 | 64 | M | 1 | 1833451010 | A | 40 | 19 |
| 25 | R4457 | LXS32 | 66 | M | 1 | 1953348019 | E | 55 | 1 |
| 26 | R4458 | LXS32 | 70 | F | 1 | 1953348019 | F | 55 | 1 |
| 27 | R3903H | LXS36 | 70 | F | 1 | 1870382055 | E | 50 | 13 |
| 28 | R3924H | LXS36 | 65 | M | 1 | 1870382090 | F | 51 | 14 |
| 29 | R3756H | LXS39 | 67 | M | 1 | 1725572045 | D | 4 | 6 |
| 30 | R3927H | LXS42 | 65 | M | 1 | 1825397042 | D | 27 | 8 |
| 31 | R3734H | LXS42 | 71 | F | 1 | 1825397077 | A | 31 | 12 |
| 32 | R3873H | LXS43 | 64 | M | 1 | 1736925163 | B | 16 | 10 |
| 33 | R3874H | LXS43 | 64 | M | 1 | 1825397073 | C | 29 | 16 |
| 34 | R3878H | LXS46 | 64 | M | 1 | 1825397073 | D | 29 | 16 |
| 35 | R3943H | LXS46 | 63 | F | 1 | 1833451001 | C | 38 | 17 |
| 36 | R3782H | LXS50 | 70 | M | 1 | 1825397032 | C | 22 | 15 |
| 37 | R3796H | LXS50 | 66 | F | 1 | 1825397073 | E | 29 | 16 |
| 38 | R3868H | LXS51 | 67 | F | 1 | 1735640066 | B | 7 | 14 |
| 39 | R3869H | LXS51 | 67 | M | 1 | 1825397080 | B | 33 | 16 |
| 40 | R3920H | LXS54 | 73 | M | 1 | 1736925163 | F | 16 | 10 |
| 41 | R3822H | LXS66 | 66 | M | 1 | 1736925120 | D | 9 | 9 |
| 42 | R3858H | LXS66 | 61 | F | 1 | 1833451001 | E | 38 | 17 |
| 43 | R3759H | LXS78 | 68 | M | 1 | 1825397077 | E | 31 | 12 |
| 44 | R3764H | LXS78 | 70 | F | 1 | 1825397100 | D | 35 | 17 |
| 45 | R3843H | LXS80 | 62 | M | 1 | 1735640066 | E | 7 | 14 |
| 46 | R3842H | LXS80 | 62 | F | 1 | 1870382102 | C | 52 | 13 |
| 47 | R3719H | LXS88 | 71 | M | 1 | 1716756046 | C | 3 | 6 |
| 48 | R3723H | LXS88 | 72 | F | 1 | 1825397036 | F | 25 | 15 |
| 49 | R4464 | LXS88 | 61 | F | 1 | 1848071023 | E | 48 | 3 |
| 50 | R3907H | LXS90 | 72 | F | 1 | 1825397080 | F | 33 | 16 |
| 51 | R3908H | LXS90 | 72 | M | 1 | 1825397108 | B | 36 | 17 |
| 52 | R3696H | LXS92 | 72 | F | 1 | 1736925164 | E | 17 | 5 |
| 53 | R3689H | LXS92 | 68 | F | 1 | 1833451012 | C | 41 | 19 |
| 54 | R3725H | LXS97 | 71 | M | 1 | 1736925293 | B | 18 | 10 |
| 55 | R3912H | LXS97 | 70 | F | 1 | 1825397018 | F | 20 | 11 |
| 56 | R3882H | LXS98 | 64 | F | 1 | 1825397047 | A | 28 | 11 |
| 57 | R3883H | LXS98 | 64 | F | 1 | 1870382102 | F | 52 | 13 |
| 58 | R3897H | LXS99 | 67 | M | 1 | 1735640068 | C | 8 | 14 |
| 59 | R3931H | LXS99 | 65 | F | 1 | 1833451018 | F | 43 | 18 |
| 60 | R3749H | LXS100 | 75 | M | 1 | 1714451039 | B | 2 | 6 |
| 61 | R3788H | LXS100 | 66 | F | 1 | 1736925293 | D | 18 | 10 |
| 62 | R3891H | LXS103 | 67 | M | 1 | 1825397081 | C | 34 | 16 |
| 63 | R3840H | LXS103 | 62 | F | 1 | 1870382129 | C | 53 | 13 |
| 64 | R3893H | LXS110 | 67 | M | 1 | 1825397081 | D | 34 | 16 |
| 65 | R3847H | LXS110 | 60 | M | 1 | 1870382129 | D | 53 | 13 |
| 66 | R3698H | LXS122 | 66 | F | 1 | 1714451039 | C | 2 | 6 |
| 67 | R3684H | LXS122 | 66 | F | 1 | 1833451008 | A | 39 | 4 |
| 68 | R4447 | LXS122 | 67 | M | 1 | 1848071028 | D | 49 | 2 |
| 69 | R3772H | LXS123 | 74 | F | 1 | 1825397035 | E | 24 | 15 |
| 70 | R3744H | LXS123 | 73 | M | 1 | 1825397076 | C | 30 | 12 |
| 71 | R3800H | LXS123 | 65 | M | 1 | 1825397078 | A | 32 | 12 |
Downloading all data:
All data links (right-most column above) will be made active as soon as the global analysis of these data has been accepted for publication. Please see text on Data Sharing Policies, and Conditions and Limitations, and Contacts. Following publication, download a summary text file or Excel file of data. Please contact Lu Lu or RW Williams if you have questions about these data.
About the array platform:
Illumina Sentrix Mouse-6.1 BeadArray Platform (ILM6v1.1): The Mouse6.1 array consists of 46,643 unique probe sequences, each 50 nucleotides in length, that have been arrayed on glass slides using a novel bead technology.
ANNOTATION: In summer of 2008, Xusheng Wang and Robert W. Williams reannotated the Illumina Mouse-6.1 array content. This new annotation is now incorporated into GeneNetwork. For 46643 probes on the Mouse 6.1 array platform (including control probes) we have identified 40183 NCBI Entrez Gene IDs; 22527 matched human Gene IDs; 11657 matched rat Gene IDs; 40983 NCBI HomoloGene IDs; and 22174 OMIM IDs.
Position data for the 50-mer Illumina Mouse-6 array were initially downloaded from Sanger at http://www.sanger.ac.uk/Users/avc/Illumina/Mouse-6_V1.gff.gz but we then updated all positions by BLAT analysis from mm6 positions to mm8 positions (Hongqiang Li).
About data processing:
This data set uses the standard Rank Invariant method developed by Illumina and described in their BeadStation Studio documentation.
Sex of the samples was tested and validated using sex-specific probe set: Xist probe ILM104280446.

Legend: Checking that the sex of samples were labeled correctly was done using Xist expression measured by probe ILM106520068. In this bar chart the expression of Xist is very low in ILS and in six of the LXS strains: LXS43, 110, 54, 78, 39, and 25. This is because both arrays are male samples rather than 1 male and 1 female sample. In contrast LXS19 and LXS92 have very high expression. LXS19 data is from a single female pool (no error bar) whereas LXS92 is from tow female pools.
Data source acknowledgment:
Data were generated with funds to Lu Lu from the NIAAA INIA program. Informatics support provided by NIH NIAAA INIA grants to RWW and LL.
Lu Lu, M.D.
Grant Support: NIH U01AA13499, U24AA13513 (Lu Lu, PI)
About this text file:
This text files was initially entered by Robert W. Williams, Oct 20, 2008. The data set was entered by Arthur Centeno and Lu Lu, Oct 15, 2008. Please contact Dr. Lu Lu regarding these expression data at lulu@utmem.edu. Updated by Robert W. Williams, Oct 21, 2008. Updated by Lu Lu on Oct 22, 2008.
|
|
|
|